All News
Varshney Named Dean of the College of Computer, Mathematical, and Natural Sciences

Computer scientist Amitabh Varshney has been named dean of the University of Maryland’s College of Computer, Mathematical, and Natural Sciences (CMNS), effective March 1, 2018.
Varshney is a professor of computer science at UMD and director of the University of Maryland Institute for Advanced Computer Studies (UMIACS). He recently completed a one-year term as the university’s interim vice president for research.
In his new role, Varshney will lead the university’s largest college, which educates more than 9,000 students each year in its undergraduate and graduate programs. He will also provide strategic leadership to expand the magnitude and impact of the college’s education and research programs, which have a combined annual budget of $250 million.
“I am deeply honored to be chosen to lead this college and bring its excellence in research and academics to new levels of visibility and recognition,” Varshney said. “I look forward to championing the efforts of our talented faculty, staff and students who are advancing new frontiers of scientific knowledge every day.”
Varshney’s appointment comes after a six-month national search.
“[Dr. Varshney] has a wealth of knowledge in research and interdisciplinary work,” said Mary Ann Rankin, UMD’s senior vice president and provost. “His clear vision for the college and natural leadership style will allow CMNS to continue to be recognized nationally and globally.”
Go here to read the full CMNS news release.
Cummings Named Interim Director of CBCB

A noted expert in computational biology and molecular evolutionary genetics has just been named interim director of the Center for Bioinformatics and Computational Biology (CBCB).
Michael Cummings, a professor of biology with an appointment in the University of Maryland Institute for Advanced Computer Studies (UMIACS), assumed leadership of CBCB on January 22.
He succeeds Eytan Ruppin, who started a new position as chief scientist of a cancer data science lab at the National Cancer Institute.
As interim director of CBCB, Cummings will help guide a diverse community of faculty, research scientists, postdocs, students and staff who are focused on research and scholarship arising from the genome revolution.
"I am privileged to collaborate with such a talented group of people, all of whom share a common interest of using powerful computational resources to gain a better understanding of how life works," says Cummings. "The work we do in CBCB—whether stemming the spread of cholera or other infectious diseases, helping curtail the use of harmful tobacco products, or analyzing synthetic DNA for possible biological threats—has a positive impact on the world we inhabit."
Cummings own research is focused on innovative experimental design and data analysis methods. This includes examining DNA sequence data for evolutionary inference, sampling of genetic diversity in relationship to conservation reserve design, hypothesis testing using tree-based statistical models, genetic differentiation, and more.
Cummings—working closely with doctoral student Daniel Ayres—was recognized last year by technology leader NVIDIA with a Global Impact Award for the development of innovative software that provides a rapid analysis of biological sequence data.
Known as BEAGLE, for Broad-platform Evolutionary Analysis General Likelihood Evaluator, the open-source software is an essential component in the software workflow of many scientists studying the evolutionary history of organisms, including the viruses that cause AIDS, influenza and Ebola.
Cummings came to the University of Maryland in 2003 as a visiting associate professor. He accepted full-time appointments in the Department of Biology and UMIACS in 2005.
Cummings received his doctorate in organismic and evolutionary biology from Harvard University in 1992, and completed postdoctoral research at the University of California, Berkeley, as an Alfred P. Sloan Foundation Postdoctoral Fellow. He also did postdoctoral work at the University of California, Riverside.
Hannenhalli and CBCB Doctoral Student Release New Study Examining Protein Networks and Breast Cancer
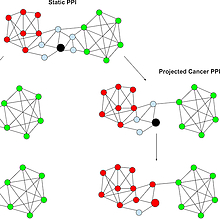
Researchers in the Center for Bioinformatics and Computational Biology (CBCB) have published a new study on how any given gene may perform a different function in breast cancer cells—as opposed to healthy cells—due to changes in networks of interacting proteins.
Previous research has shown that a protein produced by a single gene can potentially have different functions in a cell depending on the proteins with which it interacts.
Building on this concept, Sridhar Hannenhalli, a professor of cell biology and molecular genetics, and Sushant Patkar, a second-year doctoral student in computer science, hypothesized that alterations in protein interaction networks in breast cancer cells may change the function of individual genes.
To test this idea, they analyzed protein expression in 1,047 breast cancer tumors and 110 healthy breast tissue samples, using data from The Cancer Genome Atlas project.
Paktar was lead author on the research paper, which was just published in PLOS Computational Biology. Assaf Magen, a fourth-year doctoral student in CBCB, and Roded Sharan, a professor of computer science and a member of the Edmond J. Safra Center for Bioinformatics at Tel Aviv University, also contributed to the paper.
Go here to read the full news release.
Colwell Receives Prestigious Appointment to France’s Legion of Honor

Rita Colwell, a Distinguished University Professor in the University of Maryland Institute for Advanced Computer Studies (UMIACS), has been awarded the rank of Chevalier (Knight) in the Legion of Honor by decree of the President of the French Republic. Colwell was presented the Insignia of Chevalier de la Légion d’Honneur by Gérard Araud, Ambassador of France to the United States, in a ceremony on November 20 at the ambassador’s residence in Washington, D.C.
France's Legion of Honor, which was created by Napoleon Bonaparte in 1802, rewards both military and civilian merit. According to the Légion d'Honneur website: “The Legion of Honor brings together, around the criterion of personal merit in the service of the nation [France], famous personalities as well as citizens unknown to the broader public.”
“It gives me great pleasure to extend my warmest congratulations to you on being recently appointed to the rank of Chevalier in the Legion of Honor by decree of the President of the French Republic,” wrote Ambassador Araud in the official award letter.
“This high honor reflects France’s profound gratitude as expressed by the President of the French Republic and acknowledges your exemplary personal commitment to trans-Atlantic collaboration with several French universities in the fields of environment, water and global infectious diseases,” continued Araud, who also cited Colwell’s major role in the promotion of science and in inspiring many students around the world to pursue scientific careers.
Colwell's career bridges many areas, including microbiology, ecology, infectious disease, public health and computer and satellite technology. She continues to be a world leader in the fight against water-borne diseases, particularly cholera, and in bioinformatics, especially for understanding microbiomes and applying this knowledge to human health and to the diagnosis and treatment of disease.
Colwell’s many achievements and firsts include:
• establishing the taxonomy of vibrio bacteria, which includes Vibrio cholerae and identifying a previously unknown survival strategy of dormant vibrio cells;
• showing how climate change has expanded the habitat range of vibrios, and the occurrence of cholera;
• helping prevent the spread of cholera in developing countries by discovering and demonstrating an effective way to use the sari, the traditional dress of women on the Indian subcontinent, as a filter to remove vibrio-carrying plankton from drinking water drawn from ponds, rivers and other surface waters;
• receiving a dozen U.S. patents, most involving computational biology;
• founding the company CosmosID, which uses next-generation DNA sequencing to advance new discoveries in microbiome research; and
• leading numerous science organizations, including the National Science Foundation (NSF) from 1998–2004 as NSF’s first woman director.
The Legion of Honor is the latest of Colwell’s many recognitions and awards, which also include the 2017 International Prize for Biology, recognized as one of the most prestigious honors for a natural scientist; the 2006 U.S. National Medal of Science; the 2010 Stockholm Water Prize; “The Order of the Rising Sun, Gold and Silver Star,” awarded by the Emperor of Japan; and membership in the U.S. National Academy of Sciences, Royal Society of Canada, Swedish Royal Academy of Science, Irish Royal Academy of Science, the Bangladesh and Indian academies of Science, the American Academy of Arts and Sciences, American Philosophical Society and the National Academy of Inventors.
CHIB to Co-Host Microbiome Workshop on Biodefense and Pathogen Detection
The Center for Health-related Informatics and Bioimaging (CHIB) is co-hosting a one-day workshop next January focused on biodefense and pathogen detection.
The 2018 Winter Mid-Atlantic Microbiome Meet-up (M3), set for January 10, 2018 at the University of Maryland, is designed to create synergistic connections between academia, federal experts and others involved in pathogen detection, antibiotic resistance and biodefense matters.
Keynote speakers for the event are Tara O'Toole, executive vice president of In-Q-Tel (IQT), and Rita Colwell, a Distinguished University Professor in the University of Maryland Institute for Advanced Computer Studies.
O’Toole is currently leading B.Next, an IQT lab dedicated to exploring opportunities and risks likely to arise as a result of advances in the biological sciences and biotechnologies, with a particular focus on detecting and preventing biological attacks.
Colwell is recognized as an international leader in the field of bioinformatics—most notably in understanding microbiomes and the application of this knowledge to human health and the diagnosis and treatment of disease. She is founder and chairman of CosmosID Inc., a microbial genomics company focused on molecular diagnostics of human pathogens and antimicrobial resistance.
Other featured speakers are Nicholas Bergman, a senior principal investigator in genomics at the National Biodefense Analysis and Countermeasures Center, and Stephanie Rogers, a deputy director of B.Next.
This is the second Mid-Atlantic Microbiome Meet-up held on the Maryland campus. The inaugural M3 in Fall 2016 brought together more than 60 researchers and scientists who discussed topics such as human skin microbiomes and tracking the spread of hospital infections.
“M3 has served as a conduit for the Mid-Atlantic scientific community interested in microbiome research, enabling researchers at all levels to coalesce around the main challenges in the field,” says Todd Treangen, the assistant director of CHIB who is coordinating much of the M3 activities.
To register for the January 2018 workshop, go here.
University of Maryland Joins International Phytobiomes Alliance

The International Alliance for Phytobiomes Research is pleased to announce that the University of Maryland (UMD) has joined the organization as a sponsoring partner.
The Alliance is an international, nonprofit consortium of academic institutions, private companies, and government agencies. Launched in 2016, it is building a foundation for exploitation of the phytobiome for food security. “Phytobiomes” refers to a plant growing within a particular biome, enabling a holistic view of the production of crops, trees, and grasslands that includes the plant itself, all micro- and macro-organisms living in, on, or around the plant—such as microbes, animals, insects, and other plants—as well as the environment, which includes soil, air, water, weather, and climate.
The University of Maryland is one of the nation’s preeminent public research universities. Researchers in the University of Maryland Institute for Advanced Computer Studies (UMIACS), will bring extensive knowledge of genomic sequencing and bioinformatics to the Alliance, allowing for a detailed analysis of soil, water and air samples, as well as the organisms and other elements that inhabit those environments.
The university, located just outside of Washington, D.C., also has close research and educational ties to U.S. government labs and agencies like the U.S. Department of Agriculture, U.S. Food and Drug Administration, the National Institutes of Health, earth and climate specialists and the nearby NASA Goddard Space Flight Center, and more.
“The Phytobiomes Alliance proposes a holistic approach to solving one of the critical challenges facing our world—the sustainable production of food for the growing world population,” says Mihai Pop, a computational biologist at UMD who will be leading collaborative efforts with the Alliance. “We expect to contribute by bringing to bear unique expertise in machine learning and data science, high-performance computing, and metagenomics.”
Representatives of Phytobiomes Alliance sponsors serve on the organization’s coordinating committee, which identifies research projects, resource and technology gaps, establishes priorities, and develops strategic plans to achieve the Alliance’s goals.
“The University of Maryland brings to the Alliance a broad range of expertise in agricultural sciences including genetics, genomics, breeding, metagenomics, earth, climate, bioinformatics, and new technologies in high-performance computing,” says Kellye Eversole, the Phytobiomes Alliance executive director. “Mihai Pop’s background, experience, and knowledge will be extremely beneficial to the Alliance as we develop not only databases of disparate biological and geophysical information but also predictive and prescriptive analytics that could help growers increase the sustainable production of food, feed, and fiber.”
Over the next decades, the United Nations predicts that the world population will grow by 83 million every year, to reach 9.8 billion by 2050 and 11.2 billion by 2100. Producing enough food for this growing population—in a sustainable way, while preserving biodiversity and natural resources—requires a major paradigm shift in agricultural production of food, feed and fiber. By establishing a foundation of knowledge on how phytobiome components interact and affect each other, the Phytobiomes Alliance aims at addressing these challenges.
By 2050, the Alliance envisions that all farmers, ranchers, growers, and foresters will have at their disposal predictive and prescriptive tools that empower them to choose the best combination of crops, management practices, and inputs for a specific field in a given year.
About the Phytobiomes Alliance The Phytobiomes Alliance is an international, nonprofit alliance of industry, academic, and governmental partners created in 2016. The goal of the Alliance is to understand, predict and control emergent phenotypes for sustainable production of food, feed and fiber on any given farm. The Phytobiomes Alliance is sponsored by Bayer CropScience, Eversole Associates, Monsanto, The Climate Corporation, INRA, Indigo, the Noble Research Institute, NewLeaf Symbiotics, Colorado State University, the University of Maryland, Penn State College of Agricultural Sciences, the American Phytopathological Society, the University of Nebraska-Lincoln, and BioConsortia.
About the University of Maryland The University of Maryland, the state’s flagship institution of higher education, is one of the nation’s preeminent public research universities. A global leader in scientific discovery, entrepreneurship and innovation, UMD is home to nearly 38,000 students, 9,500 faculty and staff, and 250 academic programs. The institution secures more than $500M in external research funding each year.
About the Institute for Advanced Computer Studies The University of Maryland Institute for Advanced Computer Studies (UMIACS) provides a unique multidisciplinary computing environment to address some of today’s most pressing scientific and societal challenges in areas like cybersecurity, bioinformatics, computer vision, data science, and quantum information science.
CBCB Doctoral Student Uses Powerful Computing to Unlock the Mysteries of Microbes
Nidhi Shah loves a good challenge and is inquisitive by nature, so it’s only fitting that she’s involved in an expanding scientific field where curiosity and determination are essential: metagenomics.
Shah, a second-year computer science doctoral student in the Center for Bioinformatics and Computational Biology (CBCB), is developing algorithms and software tools to unlock the mysteries of microbes, the tiny microorganisms that make up the majority of our planet’s living material.
Until the last decade or so, there were many unanswered questions surrounding microbes, mostly due to the fact that only a small percentage of them could be isolated and studied in a controlled lab environment.
Now, with advances in high-throughput DNA sequencing and other tools, scientists are able to take an environmental sample—whether from a hydrothermal vent or the gut of a sheep—sequence the DNA in it, and then piece together the genomes of any microbes that are present.
One of the biggest challenges in metagenomics is to annotate, or identify, which microorganism or gene that a specific DNA sequence originates from, which is where the research by Shah and other computational biologists comes into play.
“Annotating the sequences found in these samples is complicated work, but it’s important because it helps us not only understand the diversity of our environment, but also lets us better characterize a microbe’s functional capabilities,” Shah says. “This, in turn, can help us understand and have a positive impact on human health, agricultural yield, the environment, and more, by exploring these types of microbial dynamics.”
Helping Shah in her work are the vast computing resources available in CBCB, one the dozen-plus labs and centers in the University of Maryland Institute for Advanced Computer Studies (UMIACS).
“I use UMIACS’ computational resources extensively,” she says. “It’s practically impossible to run and test our algorithms and ideas for large-scale study samples on our [desktop] computers. So being able to quickly migrate workloads to the large-scale UMIACS data center lets me quickly make informed decisions about the capabilities of our sampling methods.”Shah says that prior to coming to the University of Maryland, she had very little experience with computational biology. As an undergraduate majoring in information technology at the Veermata Jijabai Technological Institute in Mumbai, she studied computer science theory. When it came time to apply to graduate school, Shah searched for something that would be a “good mix” of both theory and application.
The cutting-edge research in metagenomics underway in CBCB seemed to be a good pathway to explore, she says.
At Maryland, Shah is working closely with her adviser, Mihai Pop, a professor of computer science who has a joint appointment in UMIACS.
Pop, an expert in metagenomics, says that Shah has made tremendous progress in her research over the past year, publishing her initial results in the recent Workshop on Algorithms in Bioinformatics (WABI 2017) held in Boston.
“We are already using the new approaches she has developed to study microbial communities from the guts of children from the developing world, as well as to study the impact of brain injury on the gastrointestinal track of mice,” Pop says. “Nidhi has strong technical skills, and an amazingly positive and thoughtful attitude—attributes that foretell an outstanding scientific career in graduate school and beyond.”
Although Shah doesn’t know exactly where her path will lead once her doctoral program is complete, she says that working in CBCB is not only teaching her computational biology, but is also providing the academic tools needed to become an independent researcher.
“Whatever field I end up working in, I want to be able to ask the right questions, and have the intuitive knowledge needed to answer those questions effectively,” she says. “Equally important is being able to communicate one’s research findings to the public. Working with all of the amazing research scientists, postdocs and other graduate students in CBCB is definitely helping me master these important skills.”
—Story by Melissa BrachfeldPop Lends Expertise to International Competition for Metagenomics Software

Communities of bacteria live everywhere: inside our bodies, on our bodies and all around us. The human gut alone contains hundreds of species of bacteria that help digest food and provide nutrients, but can also make us sick. To learn more about these groups of bacteria and how they impact our lives, scientists need to study them. But this task poses challenges, because taking the bacteria into the laboratory is either impossible or would disrupt the biological processes the scientists wish to study.
To bypass these difficulties, scientists have turned to the field of metagenomics. In metagenomics, researchers use algorithms to piece together DNA from an environmental sample to determine the type and role of bacteria present. Unlike established fields such as chemistry, where researchers evaluate their results against a set of known standards, metagenomics is a relatively young field that lacks such benchmarks.
Mihai Pop, a professor of computer science with a joint appointment in the University of Maryland Institute for Advanced Computer Studies, recently helped judge an international challenge called the Critical Assessment of Metagenome Interpretation (CAMI), which benchmarked metagenomics software. The results were published in the journal Nature Methods on October 2, 2017.
“There’s no one algorithm that we can say is the best at everything,” said Pop, who is also co-director of the Center for Health-related Informatics and Bioimaging at UMD. “What we found was that one tool does better in one context, but another does better in another context. It is important for researchers to know that they need to choose software based on the specific questions they are trying to answer.”
The study’s results were not surprising to Pop, because of the many challenges metagenomics software developers face. First, DNA analysis is challenging in metagenomics because the recovered DNA often comes from the field, not a tightly controlled laboratory environment. In addition, DNA from many organisms—some of which may not have known genomes—mingle together in a sample, making it difficult to correctly assemble, or piece together, individual genomes. Moreover, DNA degrades in harsh environments.
“I like to think of metagenomics as a new type of microscope,” Pop said. “In the old days, you would use a microscope to study bacteria. Now we have a much more powerful microscope, which is DNA sequencing coupled with advanced algorithms. Metagenomics holds the promise of helping us understand what bacteria do in the world. But first we need to tune that microscope.”
CAMI’s leader invited Pop to help evaluate the submissions by challenge participants because of his expertise in genome and metagenome assembly. In 2009, Pop helped publish Bowtie, one of the most commonly used software packages for assembling genomes. More recently, he collaborated with the University of Maryland School of Medicine to analyze hundreds of thousands of gene sequences as part of the largest, most comprehensive study of childhood diarrheal diseases ever conducted in developing countries.
“We uncovered new, unknown bacteria that cause diarrheal diseases, and we also found interactions between bacteria that might worsen or improve illness,” Pop said. “I feel that’s one of the most impactful projects I’ve done using metagenomics.”
For the competition, CAMI researchers combined approximately 700 microbial genomes and 600 viral genomes with other DNA sources and simulated how such a collection of DNA might appear in the field. The participants’ task was to reconstruct and analyze the genomes of the simulated DNA pool.
CAMI researchers scored the participants’ submissions in three areas: how well they assembled the fragmented genomes; how well they “binned,” or organized, DNA fragments into related groups to determine the families of organisms in the mixture; and how well they “profiled,” or reconstructed, the identity and relative abundance of the organisms present in the mixture. Pop contributed metrics and software for evaluating the submitted assembled genomes.
Nineteen teams submitted 215 entries using six genome assemblers, nine binners and 10 profilers to tackle this challenge.
The results showed that for assembly, algorithms that pieced together a genome using different lengths of smaller DNA fragments outperformed those that used DNA fragments of a fixed length. However, no assemblers did well at picking apart different, yet similar genomes.
For the binning task, the researchers found tradeoffs in how accurately the software programs identified the group to which a particular DNA fragment belonged, versus how many DNA fragments the software assigned to any groups. This result suggests that researchers need to choose their binning software based on whether accuracy or coverage is more important. In addition, the performance of all binning algorithms decreased when samples included multiple related genomes.
In profiling, software either recovered the relative abundance of bacteria in the sample better or detected organisms better, even at very low quantities. However, the latter algorithms identified the wrong organism more often.
Going forward, Pop said the CAMI group will continue to run new challenges with different data sets and new evaluations aimed at more specific aspects of software performance. Pop is excited to see scientists use the benchmarks to address research questions in the laboratory and the clinic.
“The field of metagenomics needs standards to ensure that results are correct, well validated and follow best practices,” Pop said. “For instance, if a doctor is going to stage an intervention based on results from metagenomic software, it’s essential that those results be correct. Our work provides a roadmap for choosing appropriate software.”
—Story by Irene Ying
This work was led by Alice McHardy of the Department for Computational Biology of Infection Research at the Helmholtz Centre for Infection Research and the Braunschweig Integrated Centre of Systems Biology in Braunschweig, Germany.
This work was supported by an Engineering and Physical Sciences Research Council Grant (Award No. EP/K032208/1), a U.S. Department of Energy contract (Award No. DEAC02-05CH11231) and the Cluster of Excellence on Plant Sciences program funded by the Deutsche Forschungsgemeinschaft. The content of this article does not necessarily reflect the views of these organizations.
UMIACS Partners with Fraunhofer, Signature Science on DNA Screening Technologies

Computational biologists in the University of Maryland Institute for Advanced Computer Studies (UMIACS) are collaborating with other experts to develop new approaches and tools for screening DNA sequences that might accidently—or intentionally—be altered, resulting in a biological threat.
Mihai Pop (left in photo), a professor of computer science and interim director of UMIACS, and Todd Treangen (right), an assistant research scientist in UMIACS, are working closely with the Fraunhofer Center for Experimental Software Engineering and Signature Science LLC on next-generation computational and bioinformatics tools that can quickly assess whether certain synthesized DNA strands could pose a risk.
“This is an ambitious project that will join experts in biology, bioinformatics, machine learning and software engineering,” says Pop. “The software underlying this project is extremely complex, involving an intricate chain of sophisticated software components. This chain has to work seamlessly—not only to reliably identify biological threats, but to do so under strict time and resource constraints.”
DNA synthesis has increased significantly during the past decade, Pop says, with scientists in academia and industry using automated machines to construct genes and other long strands of DNA by stringing together chemical building blocks called nucleotides in any desired sequence.
While these altered DNA strands can lead to revolutionary advances in medicine, agriculture and materials science, there is the possibility that someone could exploit synthetic DNA for harmful purposes—like creating a synthetic smallpox virus, a deadly scourge that was eradicated from nature in the late 1970s and currently exists only in a few highly secure repositories.
The Maryland researchers are subcontracted to Signature Science—a subsidiary of the Southwest Research Institute—on the Functional Genomic and Computational Assessment of Threats (Fun GCAT) program, which is managed by the Intelligence Advanced Research Projects Activity (IARPA).
For their role in Fun GCAT, researchers from UMIACS, Fraunhofer and Signature Science have identified specific tasks for each group in order to create a “bioinformatics analysis pipeline,” says Treangen, an expert in developing software that can quickly and efficiently analyze large amounts of genomic data.
Treangen is working on the project with Dan Nasko, a postdoctoral scientist, and graduate students in the Center for Bioinformatics and Computational Biology, an interdisciplinary center in UMIACS with access to powerful computing and data storage resources.
“My group will develop software modules that can provide rapid sequence and protein structure comparisons to assess the threat potential of functional elements from short DNA sequences,” he says. “The biggest challenge will be adapting current tools—and developing new tools—to perform accurate taxonomic assignment, function prediction, and threat assignment of these sequences.”
The scientists at Fraunhofer will integrate the software modules designed by Treangen’s team into a larger software infrastructure that meets regulatory standards and can be optimized for peak performance, says Adam Porter, a professor of computer science at the University of Maryland who is the executive director of Fraunhofer.
Fraunhofer will also create the visual dashboards needed for monitoring overall system performance, adds Porter.
“This is a large undertaking that requires robust proficiency in designing and integrating automated systems used for testing and validating large amounts of data very quickly,” he says. “Fraunhofer and UMIACS can provide that type of expertise in force.”
As the prime contractor for the $2.9M project, Signature Science will coordinate the work done by UMIACS, Fraunhofer and other team members.
In addition to their appointments in UMIACS, Pop and Treangen have leadership roles in the Center for Health-related Informatics and Bioimaging; Pop is co-director and Treangen is assistant director.
This effort is supported by the U.S. Army Research Office. The content of this release does not necessarily reflect the position or the policy of the Government, and no official endorsement should be inferred.
About UMIACS: The University of Maryland Institute for Advanced Computer Studies is multidisciplinary research institute with more than 80 faculty and research scientists from 11 departments and six colleges on the University of Maryland campus. Its primary mission to is foster and enable cutting-edge interdisciplinary research that is grounded in computer science and that addresses pressing scientific and societal challenges.
About Fraunhofer: The Fraunhofer Center for Experimental Software Engineering is an applied research and technology transfer organization that is affiliated with the University of Maryland. It regularly collaborates with UMD faculty, labs and centers to add Fraunhofer’s software and systems engineering skills to their basic research, so it can be applied to important scientific and societal problems in biology, health, national security and more.
About Signature Science LLC: A subsidiary of the Southwest Research Institute, Signature Science is a scientific and technical consulting firm providing multidisciplinary applied research, technology design and development, and scientific, technical and operational services to government and industry.
About IARPA: Launched in 2006, the Intelligence Advanced Research Projects Agency invests in high-risk, high-payoff research programs that address some of the most difficult scientific challenges faced by the U.S. intelligence community.
Pop and Treangen Assume Leadership Roles at CHIB

Two computational biologists at the University of Maryland have assumed leadership positions in the Center for Health-related Informatics and Bioimaging (CHIB), a unique cross-institutional collaboration that matches data-intensive biomedical problems to cutting-edge expertise in data science.
Mihai Pop, professor of computer science and interim director of the University of Maryland Institute for Advanced Computer Studies (UMIACS), is the new co-director of CHIB. Todd Treangen, a research scientist in UMIACS, is CHIB’s new assistant director.
As co-director, Pop will share senior leadership responsibilities with Owen White, a genomic expert at the University of Maryland School of Medicine.
Launched in 2013, CHIB matches computing resources at the University of Maryland, College Park with clinical and biomedical expertise at the University of Maryland, Baltimore.
Interdisciplinary teams from both institutions—with administrative support, and in some cases, seed funding from CHIB—have advanced research and education in areas like the use of new visualization technologies in healthcare, studying diarrheal pathogens in young children from low-income countries, and training graduate students in network biology.
The driving force in much of the center’s work is the use of cutting-edge computing technologies to address grand challenges in genomic research, medical information management and precision medicine—and then translate those findings to practitioners focused on improving human health.
“We want to build on the success we’ve had, and continue to develop new tools and technologies that will have a positive health impact on people in the state of Maryland and beyond,” says Pop.
A recent round of funding from MPowering the State will spur several new initiatives in CHIB.
One involves a series of MPower “summits”—joining CHIB experts with other researchers in the University of Maryland Medical System, other colleges and universities, and nearby federal scientists—in key areas like health data mobilization, personalized medicine and cancer, network analytics, and antibiotic resistance and pathogen detection.
“All of these will require input from a broad range of experts at both institutions—computer scientists, statisticians, physicians, epidemiologists, social science specialists, and more,” says Pop. “We're looking forward to these new research partnerships and projects.”
