CBCB Seminar Series
Most CBCB seminars are held from 2
p.m. until 3:15 p.m. on Thursdays
in the CBCB seminar room, 3118 at Biomolecular Sciences Building #296
Some external seminars are listed here. These, and some other
exceptions, will have a different time and/or place.
For directions to CBCB please scroll down.
2:00 p.m., Friday, April 9, 2010
Title: "Transcript Assembly and Abundance Estimation with High-Throughput RNA Sequencing"
By: Cole Trapnell,
CBCB
Venue: 3118 in CBCB (our usual venue)
Speaker information: Cole Trapnell will be presenting his PhD thesis
2:00 p.m., Thursday, April 15, 2010
RECOMB 2010 Practice Talks
Venue: 3118 Biomolecular
Sciences
directions
Title (RECOMB 2010 Practice
Talk): "Dense Subgraphs with
Restrictions and Applications to Gene Annotation Graphs"
Authors: Barna Saha, Allison
Hoch, Samir Khuller, Louiqa Raschid and Xiao-Ning Zhang
Speaker: Barna Saha,
a third year Computer Science graduate student working with Samir
Khuller on
algorithm
design and analysis.
Abstract:
We focus on finding complex annotation patterns representing
novel and interesting hypotheses from gene annotation data.
We define a generalization of the densest subgraph problem by adding
an additional distance restriction (defined by a separate metric)
to the nodes of the subgraph.
We show that while this generalization makes the problem NP-hard
for arbitrary metrics,
when the metric comes from the distance metric of a tree, or an
interval graph, the problem can be solved optimally in polynomial time.
We also show that the densest subgraph problem with a specified
subset of vertices that have to be included in the solution
can be solved optimally in polynomial time. In addition, we consider
other extensions when not just one solution needs to be found, but
we wish to list all subgraphs of almost maximum density as well.
We apply this method to a dataset of genes and their annotations
obtained from The Arabidopsis Information Resource (TAIR).
A user evaluation confirms that the patterns found in the distance
restricted densest subgraph for a dataset of photomorphogenesis genes
are indeed validated in the literature; a control dataset validates
that these are not random patterns. Interestingly, the complex
annotation patterns potentially lead to new and as yet unknown
hypotheses.
We perform experiments to determine the properties of the dense
subgraphs, as we vary parameters, including the number of genes and the
distance.
-------------
Title (RECOMB 2010 Practice
Talk): "Extracting between-pathway models from E-MAP interactions using expected graph compression"
Speaker: David Kelley
Abstract:
Genetic interactions (such as synthetic lethal interactions)
have become quantifiable on a large-scale using the epistatic miniarray
profile (E-MAP) method. An E-MAP allows the construction of a large,
weighted network of both aggravating and alleviating genetic interactions
between genes. By clustering genes into modules and establishing relationships
between those modules, we can discover compensatory pathways.
We introduce a general framework for applying greedy clustering
heuristics to probabilistic graphs.We use this framework to apply a graph
clustering method called graph summarization to an E-MAP that targets
yeast chromosome biology. This results in a new method for clustering
E-MAP data that we call Expected Graph Compression (EGC). We validate
modules and compensatory pathways using enriched Gene Ontology
annotations and a novel method based on correlated gene expression
from a comprehensive collection of expression experiments. EGC finds a
number of modules that are not found by any of the previous methods to
cluster E-MAP data. Further, EGC uncovers core submodules contained
within several previously found modules, suggesting that EGC can reveal
the finer structure of E-MAP networks.
2:00 p.m., Thursday, April 29, 2010
Title:
"Structural Assembly of Molecular Complexes Based on Residual Dipolar Couplings"
Speaker: Konstantin
Berlin, a finishing PhD student in Computer Science
Venue: 3118 Biomolecular
Sciences
directions
Scheduled Events
Upcoming:
- Sept. 8, 2011: Sean Eddy, Janelia Farm, HHMI
Past Events
Other Events
- UMCP
Bioscience Research & Technology Review Day (November 12, 2009)
- UMCP
Bioscience Research & Technology Review Day (November 12, 2008)
- UMCP
Bioscience Research & Technology Review Day (November 13, 2007)
- UMCP
Bioscience Research & Technology Review Day (November 16, 2006)
- UMCP
Bioscience Research & Technology Review Day (November 17, 2005)
- UMCP Hosts NIH Bioinformatics Course (January 19 & 20, 2005)
- UMCP
Bioscience Research & Technology Review Day (November 4, 2004)
- UMCP Hosts NIH Bioinformatics Course (January 22 & 23, 2004)
- UMCP
Bioscience Research & Technology Review Day (November 5, 2003)
- UMCP Hosts NIH Bioinformatics Course (January 15 & 16, 2003)
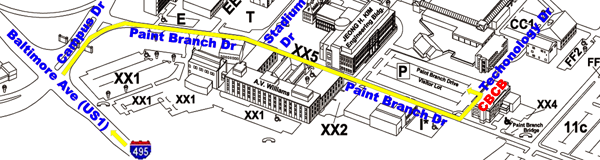
Directions
More detailed transportation options to CBCB can be found here.
From the Capital Beltway to Parking
Lot:
- take Capital Beltway (I-495) Exit 25 and turn onto Baltimore
Avenue (US Route 1) South
- go two miles south on Baltimore Ave and enter the main gate at
Campus Drive
- take the right lane into campus and make first right turn onto
Paint Branch Drive
- stop at the first stop sign then pass Stadium Drive on the left
- stop at the second stop sign then pass Parking Lot XX2 on the
right
- look for the Paint Branch Drive Visitor Lot on the left
- turn left onto Technology Drive and park in the Paint Branch
Drive Visitor Lot
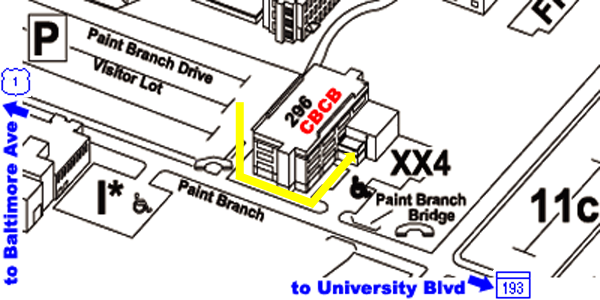
From Parking Lot to CBCB:
- the back of the Biomolecular Sciences Building #296 faces this
parking lot
- walk around to the front of the building and using the keypad
near the front door
- dial the number of one of the CBCB staff members in order to
gain entrance to the building
- CBCB is located on the third floor of the Biomolecular Sciences
Building #296
|
|