CBCB Seminar Series
For the summer 2009 Works-in-Progress series (now complete), see here.
Most CBCB seminars are held from 2
p.m. until 3:15 p.m. on Thursdays
in Room 3118 at Biomolecular Sciences Building #296
Some external seminars are listed here. These, and some other
exceptions, will have a different time and/or place.
Please see directions below.
2:00 p.m. Thursday, Oct. 8, 2009
Title: TBA.
By: Mary Schueler, National Human Genome Research
Institute, National Institutes of Health
Venue: Biomolecular
Science
Building Room 3118
2:00 p.m. Thursday, Oct. 15, 2009
Title: Computational
Techniques for Inferring Phylogenetic Relationships Using Multiple Loci.
By: Luay Nakhleh, Department of Computer Science, Rice
University
Venue: Biomolecular
Science
Building Room 3118
Abstact:
Accurate inference of phylogenetic relationships of species, and
understanding their relationships with gene trees are two central
themes in molecular and evolutionary biology. Traditionally, a species
tree is inferred by (1) sequencing a genomic region of interest from
the group of species under study, (2) reconstructing its evolutionary
history, and (3) declaring it to be the estimate of the species tree.
However, recent analyses of increasingly available multi-locus data
from various groups of organisms have demonstrated that different
genomic regions may have evolutionary histories (called “gene trees”)
that may disagree with each other, as well as with that of the
species. This observation has called into question the suitability of
the traditional approach to species tree inference. Further, when
some, or all, of these disagreements are caused by reticulate
evolutionary events, such as hybridization, then the phylogenetic
relationship of the species is more appropriately modeled by a
phylogenetic network than a tree. As a result, a new, post-genomic
paradigm has emerged, in which multiple genomic regions are analyzed
simultaneously, and their evolutionary histories are reconciled in
order to infer the evolutionary history of the species, which may not
necessarily be treelike.
In this talk, I will describe our recent work on developing
mathematical criteria and algorithmic techniques for analyzing
incongruence among gene trees, and inferring phylogenetic
relationships among species despite such incongruence. This includes
work on lineage sorting, reticulate evolution, as well as simultaneous
treatment of both.
Speaker BIO:
Luay Nakhleh is an Assistant Professor of Computer Science and
Biochemistry and Cell Biology at Rice University. He received the
B.Sc. degree from the Technion, Israel Institute of Technology, in
1996, the Master’s degree from Texas A&M University in 1998, and
the
PhD degree from the University of Texas at Austin in 2004all three
degrees in Computer Science. His research interests fall in the
general areas of computational biology and bioinformatics; in
particular, he works on computational phylogenomics and its connection
with other fields in biology. Luay has published over 50 manuscripts
on his work, supervised the dissertations of two recent PhD graduates,
and currently supervises the dissertations of 6 PhD students. Luay has
received several awards, including the Texas Excellent Teaching Award
from UT Austin in 2001, the Outstanding Dissertation Award from UT
Austin in 2005, the Roy E. Campbell Faculty Development Award from
Rice University in 2006, the DOE Early Career Award in 2006, the NSF
CAREER Award in 2009, and the Phi Beta Kappa Teaching Prize in 2009.
11:00 a.m. Thursday, Oct. 22, 2009
Title:
Finding the trees in Darwin's forest.
By: Robert K. Bradley, Massachusetts Institute of Technology
Venue: Biomolecular Science
Building Room 3118
Abstact:
TBA
Scheduled Events
- Fall, 2009: Mei-Ling Ting Lee,
Maryland School of Public Health
- Feb. 11, 2010: Sean
Eddy, HHMI Janelia Farm
- March 25, 2010: Olga Troyanskaya,
Princeton, "From genomics data integration to using
functional relationship networks to understand disease at the
molecular level"
Past Events
Other Events
Directions
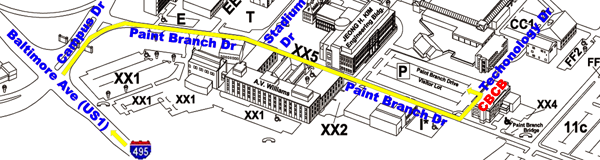
More detailed transportation options to CBCB can be found here.
From the Capital Beltway to Parking
Lot:
- take Capital Beltway (I-495) Exit 25 and turn onto Baltimore
Avenue (US Route 1) South
- go two miles south on Baltimore Ave and enter the main gate at
Campus Drive
- take the right lane into campus and make first right turn onto
Paint Branch Drive
- stop at the first stop sign then pass Stadium Drive on the left
- stop at the second stop sign then pass Parking Lot XX2 on the
right
- look for the Paint Branch Drive Visitor Lot on the left
- turn left onto Technology Drive and park in the Paint Branch
Drive Visitor Lot
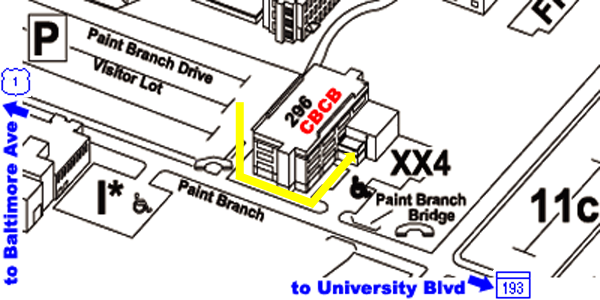
From Parking Lot to CBCB:
- the back of the Biomolecular Sciences Building #296 faces this
parking lot
- walk around to the front of the building and using the keypad
near the front door
- dial the number of one of the CBCB staff members in order to
gain entrance to the building
- CBCB is located on the third floor of the Biomolecular Sciences
Building #296
|
|