CBG Seminar Series - Spring 2011
This is the Spring 2011 archive. Please see our main page for current seminars.
Most CBCB seminars are now held jointly with the Computational Biology, Bioinformatics and Genomics Concentration Area within the Biological Sciences Graduate Program. The seminars are held from 2 p.m. until 3:15 p.m. on Thursdays in the seminar room of the Biosciences Research Building (room 1103). Starting in the fall of 2011, these seminars will be held at noon on Wednesday, alternately with seminars hosted by the Molecular and Cellular Biology graduate program. Other seminars are held in CBCB, and some external seminars of interest to CBCB are held here.
2:00 pm Thursday, Feb. 10, 2011
Title: "Chromosomal abnormalities underlying childhood brain disorders."
By: Jonathan Pevsner
Venue: 1103 Biosciences Research Bldg.
Speaker information: Jonathan Pevsner
Jonathan Pevsner is an Associate Professor in the Kennedy Krieger Institute, Dept. of Neurology and Johns Hopkins University School of Medicine, Dept. of Neuroscience. He is the author of the textbook "Bioinformatics and Functional Genomics". He will discuss methods to analyze chromosomal changes based on single nucleotide polymorphism (SNP) analyses, using a series of novel tools to determine genetic relatedness between individuals in health and disease (pevsnerlab.kennedykrieger.org). This work includes studies of patients as well as populations such as HapMap.
2:00 pm Thursday, Feb. 17, 2011
Title: "Atomistic and Coarse-Grained Simulations of DNA and Nucleosomes"
By: Garyk Papoian
Venue: 1103 Biosciences Research Bldg.
Speaker information:
Garyk Papoian is an Associate Professor in the Dept. of Chemistry and Biochemistry at the University of Maryland (since last semester). His group uses advanced computational methods to study biological processes at multiple scales, from single protein functional dynamics and chromatin folding and stability to cell-level processes, such as stochastic signal transduction and regulation of cell motility.
4:00 pm Tuesday, March 1, 2011
Rescheduled - note unusual time slot!
Title: "Estimating ultra-large phylogenies and alignments"
By: Tandy Warnow
Venue: 1103 BRB
Speaker information:
Tandy Warnow is the David Bruton, Jr. Centennial Professor in Computer Science at the University of Texas, Austin. She is spending some of her sabbatical at CBCB this spring. Her current research focuses on the development of mathematical models and algorithms for estimating evolutionary history in Biology and Historical Linguistics. The main objective is to develop methods that produce much more accurate estimations of evolutionary history than can be obtained using existing tools. Her group is distinguished by their focus on ultra-large datasets, with up to 500,000 sequences. The research is funded by the National Science Foundation via an Assembling the Tree of Life (ATOL) grant for simultaneous estimation of alignments and trees.
2:00 pm Thursday, March 10, 2011
Title: "Reconstructing tumor progression pathways from heterogeneous samples."
By: Russell Schwartz
Venue: 1103 Biosciences Research Bldg.
Speaker information:
Russell S. Schwartz is an Associate Professor at Carnegie Mellon University. His research is on the analysis of genetic variations, with specific application to inference of population subgroups and phylogenetics, and on modeling and simulation of biological systems, particularly self-assembly systems.
Abstract:
One of the major goals of current research in oncology is to understand the diversity of ways in which healthy cells can become cancerous and progressively transform into increasingly aggressive tumors. By understanding the major sequences of genetic abnormalities by which tumors develop, it is hoped that we can better characterize the common molecular causes of cancers, in turn helping us to identify subgroups of patients who will benefit from similar treatments and design effective diagnostics and therapeutics for those subgroups. Algorithms from the field of phylogenetics, or the inference of evolutionary trees, have proven a powerful method for inferring likely pathways of tumor progression. Their use has been hindered, however, by the high heterogeneity from cell-to-cell in individual tumors, making it difficult to characterize particular discrete states of progression. Our group has developed a computational strategy for attempting to characterize well-populated cell types from genomic assays of tumors, which can then be used to infer possible progression pathways. Our approach relies on “unmixing” algorithms, which use techniques derived from computational geometry to explain expression tissue-wide microarray or comparative genomic hybridization (aCGH) data as different mixtures of a common set fundamental components. We interpret these components as RNA expression or DNA copy number profiles of discrete cell types commonly produced in tumor progression. We can then apply a statistical analysis to these inferred cell types to detect significantly aberrant genes or genomic regions that can serve as markers for phylogenetic inference. Finally, we apply tree reconstruction algorithms to identify likely evolutionary pathways among the cell types. These pathways identify possible unobserved ancestral cell types and suggest specific patterns of mutations that may characterize progression between those cell types. It is hoped that this approach to tumor phylogeny inference on heterogeneous samples will prove valuable in identifying new clinically significant tumor sub-types, characterizing their causal mutations, and suggesting novel diagnostics or therapeutic targets.
2:00 pm Thursday, March 17, 2011
Title: "Optimal Experimental Design for Biological Applications"
By: Matthias Chung
Venue: 1103 Biosciences Research Bldg.
Speaker information:
Matthias Chung is an Assistant Professor at Texas State University, San Marcos. His research is on computational biology and medicine, inverse problems, numerical analysis, optimization, optimal experimental design and scientific computing.
Abstract:
Abstract:
How often, when and where should a phenomenon of interest be observed to receive reliable results? Generally, experimentalists face the dilemma between accuracy and costs of an experiment. Each experiment has its own specific challenges. However, optimization methods for inverse problems form the basic computational tool to address eminent questions of optimal experimental design. Driven by its application such as in biology, medicine, and geophysics, optimal experimental design lead to challenging optimization problems. Here, methods must address difficulties like ill-posedness, large scales, PDE or ODE constraints. We present a computational framework for the design of experiments and investigate elementary biological systems of differential equations.
2:00 pm Tuesday, April 5, 2011
Title: "Beyond counts: quantifying transcript expression with RNA-Seq and generative probabilistic models"
By: Colin Dewey
Venue: 1208 Biology Psychology Bldg.
Note the unusual time and place.
Speaker information:
Colin Dewey is Assistant Professor of Biostatistics and Medical Informatics and Computer Sciences in the Genome Center of Wisconsin at the University of Wisconsin, Madison
2:00 pm Thursday, April 7, 2011
Title:
"Chaperonin-Mediated Protein Folding"
By: Dave Thirumalai
Venue: 1103 Biosciences Research Bldg.
Speaker information:
Dave Thirumalai is Professor and Director of the Maryland Biophysics Program in the Institute for Physical Science and Technology
2:00 pm Thursday, April 14, 2011
Title: "Gene-based tests of association"
By: Joel Bader
Venue: 1103 Biosciences Research Bldg.
Speaker information:
Joel Bader is an Assistant Professor in the Department of Biomedical Engineering, Johns Hopkins University
11:00 am Friday, April 22, 2011
An informal seminar outside of the standard CBG series
Title: "Soil metagenome assembly: diverse data, diverse challenges"
By: Titus Brown
Venue: 3118 CBCB (Biomolecular Sciences Bldg.)
Abstract:
Soil contains one of the most diverse and unexplored microbial populations
on the planet. As part of the Great Prairie DOE project, the JGI has generated
several terabases (10**12) of whole metagenome shotgun sequence from a number
of sites in the midwest. Assembling this data into something meaningful has
been challenging, because it stretches current computational capacity and
algorithms. We've developed a range of approaches based on a synopsis
data structure, the Bloom filter, that lets us explore and analyze extremely
large k-mer assembly graphs, and do high-quality pre-filtering for de Bruijn
graph assemblers. This has led us to a better understanding of sequence
assembly approaches, Illumina artifacts, and assembly scaling. These results
have also enabled us to assemble very large mRNAseq data sets from emerging
model organisms.
Speaker information:
Titus Brown is an assistant professor in the departments of Computer Science and Engineering (CSE) and Microbiology and Molecular Genetics (MMG) at Michigan State University.
2:00 pm Thursday, April 28, 2011
Title: "Issues in design and analysis of microarray studies"
By: Mei-Ling Ting Lee
Venue: 1103 Biosciences Research Bldg.
Speaker information:
Mei-Ling Ting Lee is a Professor in the Department of Epidemiology and Biostatistics, University of Maryland School of Public Health
Past Events
Other Events
- UMCP
Bioscience Research & Technology Review Day (November 12, 2009)
- UMCP
Bioscience Research & Technology Review Day (November 12, 2008)
- UMCP
Bioscience Research & Technology Review Day (November 13, 2007)
- UMCP
Bioscience Research & Technology Review Day (November 16, 2006)
- UMCP
Bioscience Research & Technology Review Day (November 17, 2005)
- UMCP Hosts NIH Bioinformatics Course (January 19 & 20, 2005)
- UMCP
Bioscience Research & Technology Review Day (November 4, 2004)
- UMCP Hosts NIH Bioinformatics Course (January 22 & 23, 2004)
- UMCP
Bioscience Research & Technology Review Day (November 5, 2003)
- UMCP Hosts NIH Bioinformatics Course (January 15 & 16, 2003)
Directions
More detailed transportation options to CBCB can be found here.
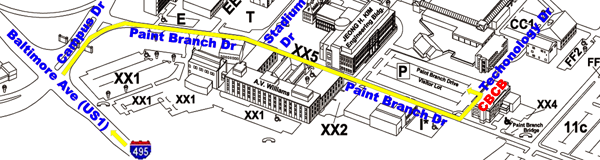
From the Capital Beltway to Parking
Lot:
- take Capital Beltway (I-495) Exit 25 and turn onto Baltimore
Avenue (US Route 1) South
- go two miles south on Baltimore Ave and enter the main gate at
Campus Drive
- take the right lane into campus and make first right turn onto
Paint Branch Drive
- stop at the first stop sign then pass Stadium Drive on the left
- stop at the second stop sign then pass Parking Lot XX2 on the
right
- look for the Paint Branch Drive Visitor Lot on the left
- turn left onto Technology Drive and park in the Paint Branch
Drive Visitor Lot
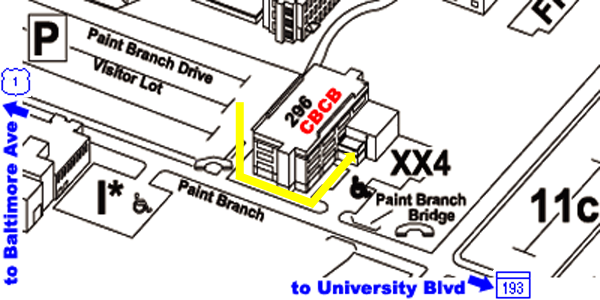
From Parking Lot to CBCB:
- the back of the Biomolecular Sciences Building #296 faces this
parking lot
- walk around to the front of the building and using the keypad
near the front door
- dial the number of one of the CBCB staff members in order to
gain entrance to the building
- CBCB is located on the third floor of the Biomolecular Sciences
Building #296
|